A first-of-its-kind database reveals how DNA mutations “destabilize” proteins and cause genetic disease.
According to RCO News Agency, A new database of half a million mutations may provide new ways to treat genetic diseases, scientists say.
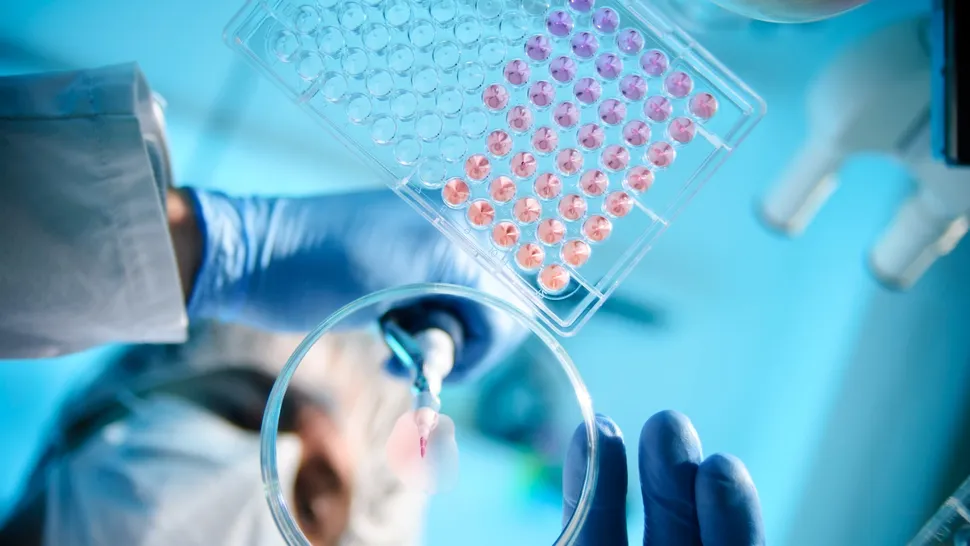
According to LiveScience, scientists have created a large database that shows how half a million different DNA mutations cause errors in human proteins. The researchers hope that this database will be used to develop new, personalized drugs that directly reverse the effects of the mutations.
The human genome contains instructions for at least 20,000 proteins that are essential for almost all physiological processes. Each protein’s structural unit, called an amino acid, is key to its function, and thus, swapping around amino acids can essentially break the protein. Missense mutations, which are a type of point mutation in which a single nucleotide change causes the genetic code to be altered in such a way that a completely different amino acid from what was supposed to be made, instead of the original amino acid, occurs in approx. 5,000 human proteins are known to cause genetic diseases such as Huntington’s disease and cystic fibrosis.
However, in many cases, it is not clear how these mutations affect the structure and function of proteins and thus cause disease. Without this understanding, it will be difficult to develop targeted therapies for genetic disorders without altering the genome itself, according to the authors of a new study published in the journal Nature on January 8.
“Depending on what happens to the protein, if you want to design a drug to cure the disease, depending on the individual mutation you’re considering, the approach will be completely different,” says Antoni Beltran, senior author of the study and one of the researchers. .
To tackle this problem, Beltran and his colleagues created a massive database that lists the effect of more than 500,000 missense mutations on the stability of 522 protein “domains,” meaning regions of proteins that are key to their function. They call the database the human “domain,” and they built it by systematically mutating proteins in the lab. Then they transferred the mutants to yeast cells and monitored the effects.
In the new study, the group specifically looked at 621 missense mutations from the database that were previously known to cause disease in humans. They found that 60% of these mutations make the affected proteins less stable. Unstable proteins are more likely to denature or lose their native state. Similar to origami, proteins must be folded in a specific way to achieve their desired shape. Improperly folded proteins can accumulate inside cells, potentially causing damage, or simply be broken down by the body, limiting the cells’ ability to function.
For example, an inherited form of cataracts, an eye disease in which the lens of the eye becomes cloudy, is caused by mutations in the genes for the beta-crystallin proteins that normally keep the lens clear. In the new study, Beltran and colleagues discovered that 72 percent of these mutations destabilize the crystallin proteins, making them more likely to aggregate and form cloudy areas in the lens.
Some missense mutations led to different changes in the proteins instead of causing instability. For example, some of the mutations behind Rett syndrome, a rare neurodevelopmental disorder, prevent a specific protein from binding to DNA. This process normally enables the protein to turn genes on and off in the brain, but in this syndrome, it goes awry.
Beltran acknowledged that although the database is the first and largest of its kind, it so far covers only 2.5 percent of known human proteins, and therefore more studies are needed to expand it.
Beltran said the team’s ultimate goal is to create a database that is useful for predicting the effect of any mutation on protein stability. Such a tool could theoretically enable scientists to develop better drugs for genetic diseases that target the proteins that go wrong and cause the disease.
end of message
RCO NEWS